FINAL REPORT: Genetic Improvement of Dry Beans for Bruchid Resistance for Southern Africa
March 15, 2024
Feed the Future Innovation Lab for Legume Systems Research
Final Report
Project Title: Genetic Improvement of Dry Beans for Bruchid Resistance for Southern Africa
Executive Summary
Common bean (Phaseolus vulgaris L.) is the most consumed grain legume in southern Africa. Besides being a major source of protein and income for many households, it is a critical component of food security. The common bean weevil (a.k.a. bean bruchid) (Acanthoscelides obtectus) (AO) is a major post-harvest pest responsible for 48-100% losses in seed quality and quantity. This project aimed to mitigate post-harvest losses of common bean and consequently, increase food security in three southern African countries Malawi, Mozambique, and Zambia. The project objectives were: (i) deploy AO resistance in preferred market classes of common bean in Malawi, Mozambique and Zambia, (ii) develop and validate molecular markers for AO resistance screening, and (iii) train the next-generation bean breeders from Malawi, Mozambique and Zambia. Three improved sister germplasm lines AO-1012-29-3-3A (AO-3A), AO-1012-29-3-1A (AO-1A), and AO1012-29-3-6B (AO-6B) that were developed co-operatively by Sokoine University of Agriculture, Oregon State University, USDA-ARS and the University of Puerto Rico were used as sources of AO resistance. The basis of AO resistance for these genotypes is the APA locus originally derived from tepary bean (P. acutifolius A. Gray). AO-3A, AO-1A and AO-6B were crossed to eight susceptible Andean genotypes selected based on the preferred market classes and developed 1,148 F5:7 breeding lines, which were evaluated for AO resistance. Sixteen breeding lines were identified as resistant, and shared with collaborators in Malawi, Mozambique, and the USA for agronomic performance, cooking time evaluation, and molecular marker validation. Three molecular markers were tested to track the bruchid resistance derived from the AO sources. The InDel marker (α-AI) showed 100% prediction accuracy for weevil resistance when the AO source is used and could be used effectively in Marker-Assisted Selection (MAS). The 16 AO-resistant lines were evaluated for cooking time using the Mattson cooker, and results showed that adding the AO resistance did not affect cooking time. In addition, several faster cooking resistant lines were identified. At least two breeding lines are currently 3 being considered for commercial release. Furthermore, the project has trained four female next-generation breeders in plant breeding at the master's level. One student from Zambia was trained at North Dakota State University, and the other three from Malawi, Mozambique, and Zambia were trained at the University of Zambia in the Plant Breeding and Seed System master’s program. This graduate training has strengthened common bean research capacity for the national breeding programs in Malawi, Mozambique, and Zambia.
Project Partners
Juan M. Osorno
Dept. of Plant Sciences, North Dakota State University (NDSU). Fargo-ND 58108.
Phil McClean
Dept. of Plant Sciences, North Dakota State University (NDSU). Fargo-ND 58108
Carlos Urrea
University of Nebraska, Panhandle Research and Extension Center, Scottsbluff NE 69391.
Kelvin Kamfwa
Dept. of Plant Sciences, University of Zambia (UNZA), Great East Road, Lusaka, Zambia.
Virginia Chisale
Dept. of Agricultural Research and Technical Services (DARS), Lilongwe, Malawi
Celestina Jochua
Instituto de Investigaciones Agrarias de Mozambique (IIAM), Maputo, Mozambique
Project Goals and Objectives
This project aimed to mitigate post-harvest losses of common bean in three southern African countries Malawi, Mozambique, and Zambia. This will increase food security in the region by providing a stable and high-quality food/protein source.
The specific objectives for the project were:
Objective 1: Develop and release weevil-resistant common bean varieties with good agronomic performance belonging to seed market classes preferred in Malawi, Mozambique, and Zambia.
Objective 2: Use existing modern breeding tools (including newly developed markers associated with bruchid resistance) to make the selection process more efficient and quicker.
Objective 3: Determination of the cooking time of improved breeding lines with bruchid resistance.
Objective 4: Training the next generation of crop improvement scientists for the region (HICD).
Overview of Activities
1. Identification of AO, and Standardization of the Weevil Screening Lab Protocol (Obj. 1)
In Zambia, both the common bean weevil (Acanthoscelides obtectus Say) (AO) and the cowpea weevil (Callosobruchus maculatus Fabricius) (CM) attack common bean. A similar situation occurs in both Malawi and Mozambique. In some other regions in Africa such as Ethiopia, a third bruchid species known as the Mexican weevil (Zabrotes subfasciatus Boheman), also cause major economic losses. Sometimes, these weevils can be found in the same storage container, specially AO along with CM. Therefore, one of the early activities conducted in the project was to correctly identify AO specimens, then build and maintain their colony for use in the project. This identification was done using a microscope complemented with a literature description of differences in physical appearance between AO and CM.
The other early activity to be undertaken by the project was to optimize and adapt the screening protocol for use in the project. Dr. James Beaver and his team at University of Puerto Rico developed the screening protocol (Kusolwa et al., 2016) that was adapted. Briefly, it involves placing ten seeds in a 250 ml plastic bottle jar, and then introducing 10 weevils. The lid of the jar is perforated for ventilation. After 65 days post infestation, the seeds are evaluated for the number of perforations, percentage of damaged seed, and the number of emerged adult weevils. Resistance is then rated on a scale of 1- 5 depending on the number of perforations in the seed, where score of 1 is resistant and 5 is susceptible. For infestation and damage to occur, the climatic conditions (temperature and humidity) in the screening room must be optimized. The screening protocol was tested, optimized, and adapted for use in the insectarium at UNZA after four test runs on susceptible and resistant bean genotypes.
2. Development of Breeding Populations and Screening for AO Resistance (Obj. 1)
Three resistant improved germplasm lines known as AO-3A (Kusolwa et al., 2016), and two additional sister lines (AO-1A, and AO-6B) that were developed cooperatively by Sokoine University of Agriculture, Oregon State University, USDAARS, and the University of Puerto Rico were used as sources of AO. The basis of AO resistance of these genotypes is the APA locus originally derived from tepary bean (P. acutifolius A. Gray). These 3 resistant lines were crossed to eight susceptible Andean genotypes. Six susceptible parents were selected from the Andean Diversity Panel (Cichy et al. 2015) based on their adaptation and agronomic performance in the southern African region. The other two susceptible parents are landraces from Zambia. Additionally, color, shape, and size of seed were important considerations in the selection so they would represent important commercial market classes in southern Africa (please refer to previous annual reports for the complete list of genotypes and crosses). The three resistant and nine susceptible parents were initially tested for AO resistance (Figure 1).
A total of 19 single reciprocal crosses were made between the three resistant and eight susceptible parents to create 19 breeding populations comprised of 1,148 F5:7 breeding lines. Of the 1,148 breeding lines, 574 were developed from the resistant parent AO -3A. These 574 were evaluated for AO resistance in the first phase of screening. The screening was conducted using a Completely Randomized Design with four replications. The first two replications of screening were conducted in the insectarium at UNZA, and the other two at the Zambia Agricultural Research Institute Station located in Northern Zambia using the lab protocol previously described. Of the 574 lines, 16 (2.8%) had higher or similar levels of resistance to the parent AO-3A (Figure 2).
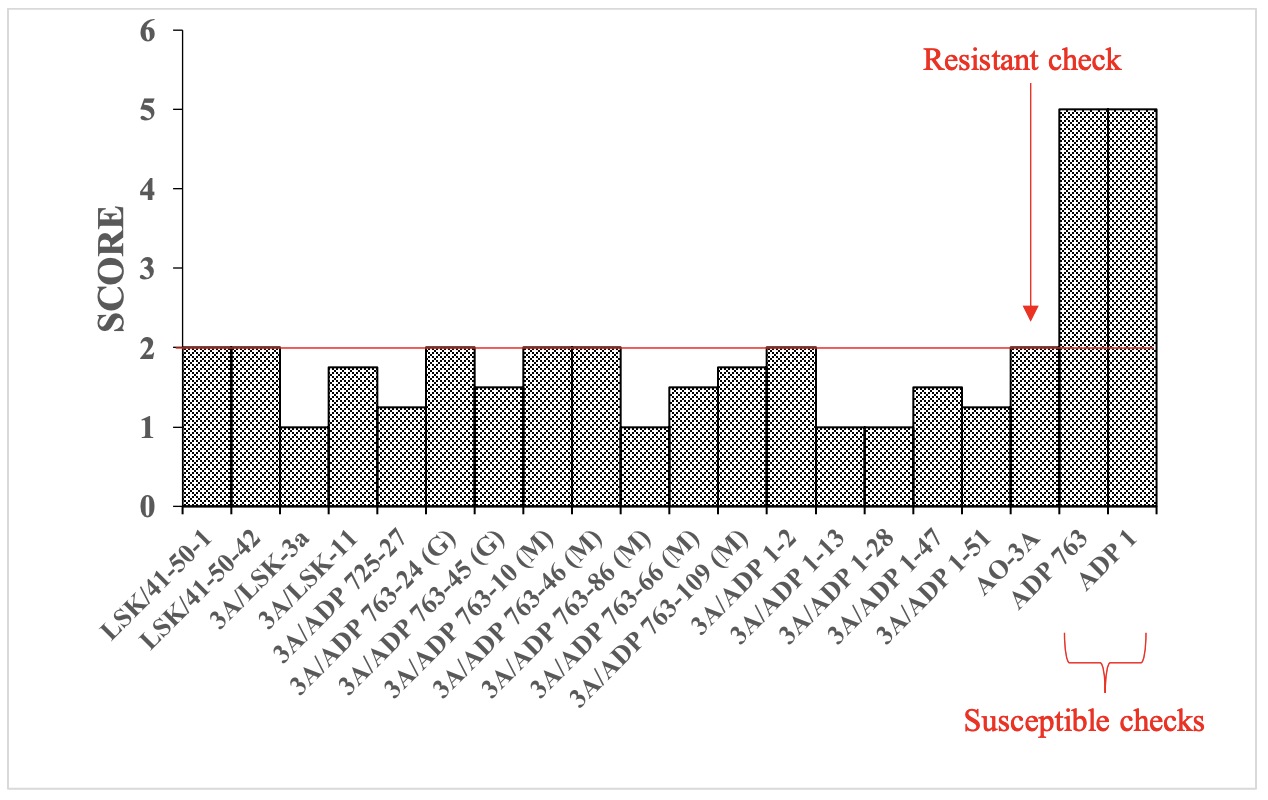
These 16 lines were in variable seed colors, shapes, and sizes, including the red mottled, which is a popular market class in Malawi, Mozambique, and Zambia (Figure 3). The second phase of screening targeted 574 lines derived from AO-1A (315 lines) and AO-6B (259 lines). Initial screening of these 574 lines from AO-1A and AO-6B has been conducted, but additional screening is currently underway for confirmation of resistance. Therefore, additional resistant lines will be confirmed in the near future.

The 16 AO resistant lines developed from AO-3A were shared with project partners in the US (NDSU and UNL), Because of low seed stocks, only a subset of 6 resistant lines were shared with Malawi (DARS), and Mozambique (IIAM) for field 9 testing and seed multiplication. These genotypes as well as the original parents and some local, resistant and susceptible checks, were tested for field agronomic performance at Malawi, Mozambique, United States (NDSU), and Zambia, tested for cooking time (UNL), screened for resistance to anthracnose disease in Zambia, and used for the validation of three molecular markers at NDSU.
3. Evaluation and Validation of Molecular Markers Associated with Bruchid Resistance for use in Marker-Assisted Selection (MAS) (Obj. 2)
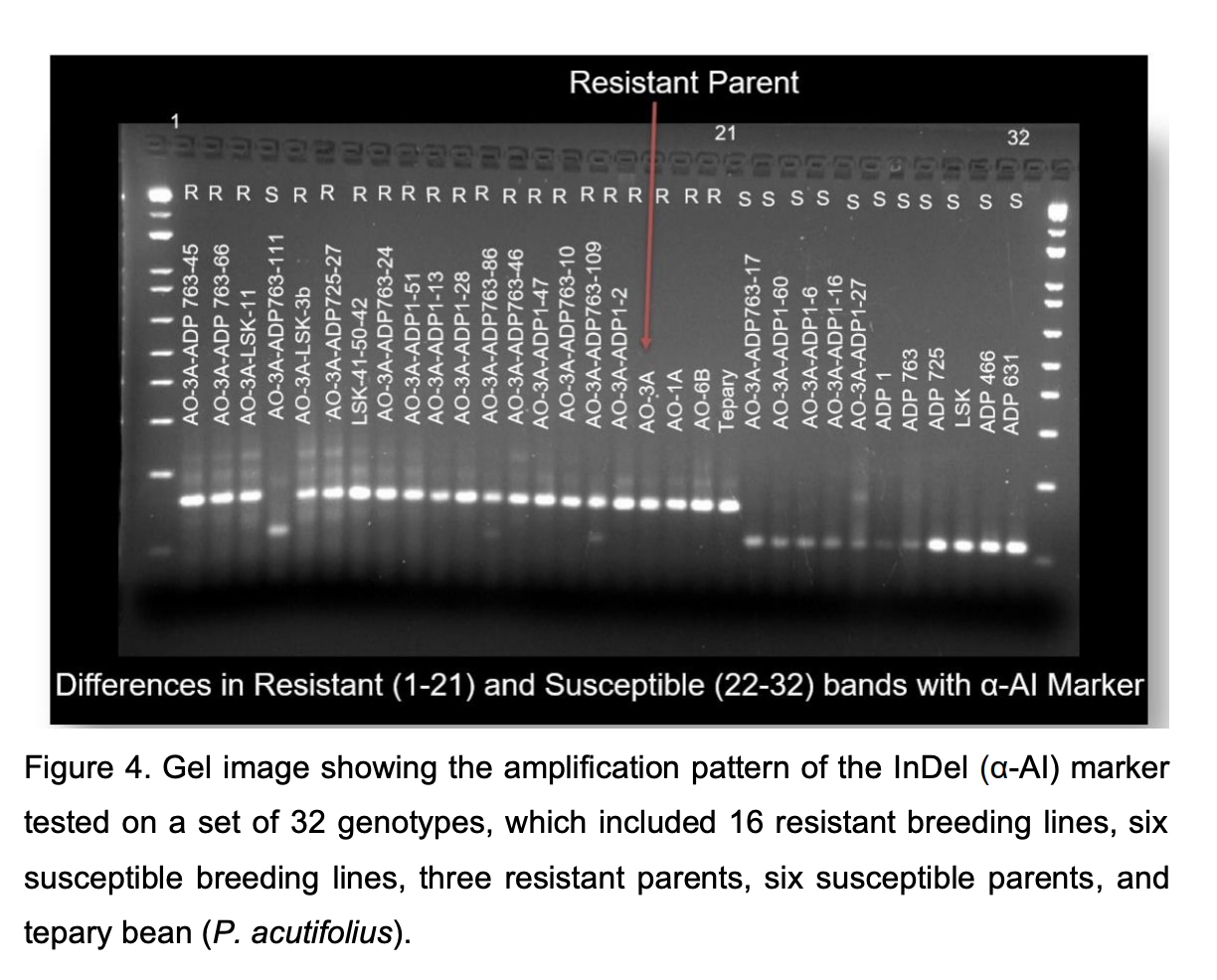
As shown before, screening for weevil resistance is time consuming, spanning about 60 days, and requires dedicated lab space with specific temperature and humidity conditions, among other factors. Molecular markers can be used to circumvent this challenge. Three molecular markers associated with bruchid resistance have been developed: an Indel marker (α-AI-1) (Mazaheri, 2018) and two SNP-KASP markers (APA, S04_46273822 and Phs, S07_5221126) (Viscarra‐Torrico et al., 2021). The project team at NDSU developed the InDel marker (α-AI) tagging the APA locus (Mazaheri, 2018) for possible use in MAS for weevil resistance. These markers were tested on a set of 32 genotypes, which included 16 resistant and six susceptible breeding lines, three resistant and six susceptible parents, and the original tepary bean (P. acutifolius) source of resistance (G40199). The UNZA project team phenotyped (for AO resistance) the 32 genotypes except the tepary accession. In addition, a set of 32 Andean bruchid resistant breeding lines developed and phenotyped by the team at the University of Puerto Rico-Mayaguez dry bean breeding program were used to test the efficiency of α-AI-1.
Once compared with the phenotypic data obtained in the lab screening, the APAS04_46273822 marker differentiated the resistant lines from the susceptible lines 79% of the times, while the Phs- S07_5221126 marker was able to distinguish the resistant and susceptible alleles only for 27% of the lines. Interestingly, the Indel marker (α-AI-1) had a 100% phenotype prediction accuracy across both sets of 10 lines (Figure 4), indicating that it can be used effectively in MAS for the AO-derived resistance.
4. Evaluation of weevil Resistant Lines for Cooking Time (Obj. 3)
Cooking time is an important end-user trait in Africa, where the percentage of canned beans is very small. Developing varieties that are weevil resistant and faster cooking is an effective strategy to mitigate post-harvest losses, increase food security, and mitigate deforestation (and child labor in some cases) caused by the use of firewood and charcoal used in cooking beans. The 16 AO-resistant breeding lines along with checks were evaluated for cooking time using a Mattson cooker in Dr. Carlos Urrea’s Lab at the University of Nebraska.
The ANOVA results showed highly significant differences among the tested breeding lines for cooking quality (Table 1). The cooking time of White Pearl (Urrea et al., 2022), a great northern cultivar (fast cooking check) averaged 33 minutes (mins) whereas AO-1012-29-3-3A (bruchid resistant parent) averaged 43 mins. The cooking time for breeding lines varied from 29 mins (AO-3A-ADP763-109) to 61.9 mins (ADP 725) with an overall mean of 43 mins (Table 1). The mean cooking time between the bruchid resistant lines and the susceptible was not statistically different, suggesting that the introgression of this bruchid resistance did not affect cooking time (Table 2). However, there were significant differences in cooking time among the 16 resistant breeding lines. The breeding line AO-3A-ADP1-13 cooked faster (35 minutes) than all other lines and the weevil-resistant check (Tables 1 and 3). Interestingly, this breeding line had shown higher AO resistance and high seed yield at the NDSU Hatton Field Trial, 2022 compared to the resistant parent AO-3A. This ideal combination of superior AO resistance, faster cooking time, and productivity shown by the breeding line AO-3A-ADP1-13 is likely to enhance its faster adoption by farmers if released as a variety.
5. Evaluation for Agronomic Performance of Weevil Resistant Lines in Malawi, Mozambique, USA, and Zambia (Obj. 1)
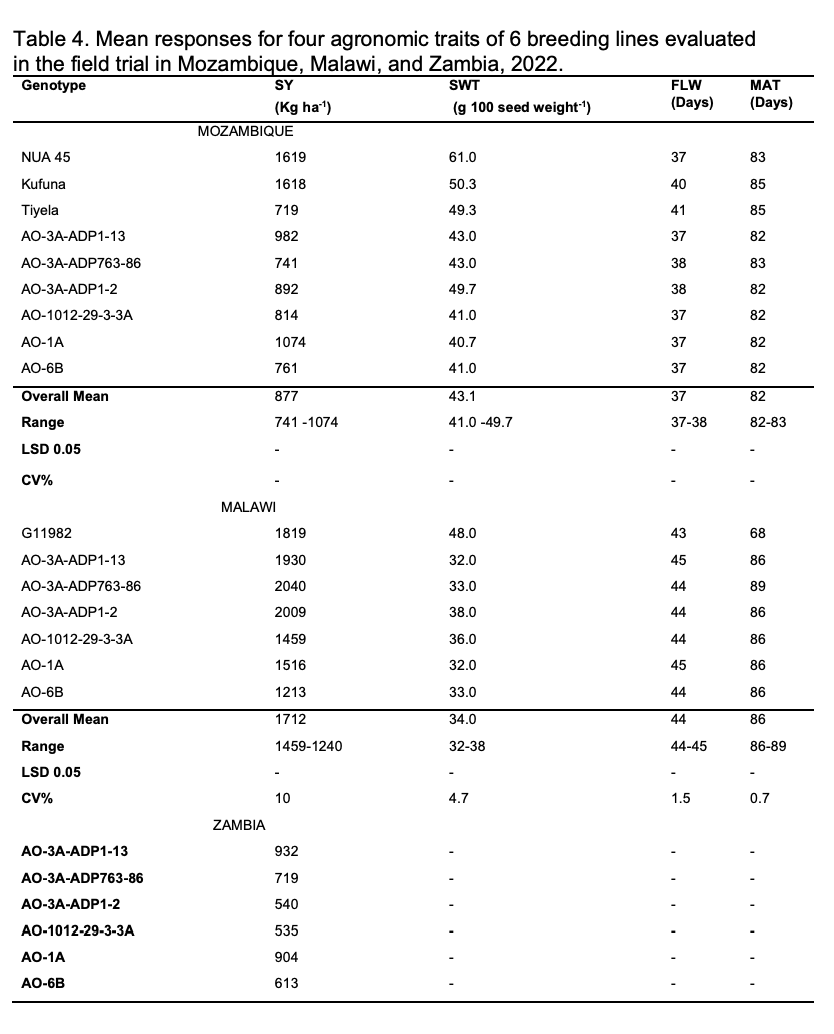
Six AO-resistant breeding lines were commonly shared with project teams in Malawi, Mozambique, and the US, where they were tested for agronomic performance in field trials. Due to low seedstocks when we started working with 14 these lines, only limited seed quantities were sent and therefore, the first year of growing these breeding lines in the three countries was to multiply the seed that was then used in the following year to conduct replicated yield trials. The results for seed yield of the 6 AO-resistant lines along with some checks at each location are shown in Table 4.
For the field trial conducted in Mozambique, the seed yield varied from 719 to 1619 kg ha-1 (Table 4). The lines that had the highest yields were NUA 45 and Kufuna, while the local check Tiyela had the lowest yield (719 kg ha-1 ). The yield of the bruchid breeding lines varied from 741 to 1074 kg ha-1 and out yielded the local check Tiyela. The highest yielding line was AO-1A (1074.5 kg ha-1 and the lowest was AO-6B (761 kg ha-1 ). In the trial conducted in Malawi, seed yield for the bruchid breeding lines varied from 2040 to 1459 kg ha-1 (Table 4). The highest yielding line was AO-3A-ADP763-86 (2040 kg ha-1 ) while the resistant parent, AO-3A (1459 kg ha-1 ) was the lowest yielder. Notably, the top three lines were AO-3A-ADP763-86 (2040 kg ha-1 ), AO-3A-ADP1-2 (2009 kg ha-1 ) and AO-3A-ADP1-13 (1930 kg ha-1 ) and most importantly, these lines out yielded the local check G11982 (1819 kg ha1 ). In the Zambian Trial (Table 4), seed yield varied from 535 to 932 kg ha-1 . The top yielding line was AO-3A-ADP1-13 (932 kg ha -1 ) and the lowest was AO-3A (resistant parent). The top yielding lines were AO-3A-ADP1-13 (932 kg ha-1 ), AO1A (904 kg ha-1 ) and AO-3A-ADP763-86 (719 kg ha-1 ).
In the same way, the ANOVA showed highly significant differences among the 30 genotypes (16 R lines plus parents and checks) for all agronomic traits measured at Hatton-ND (P<0.05, Table 5). The average seed yield ranged from 218 to 1810 kg ha-1 for the tested lines. The top yielding line was AO-3A-ADP763-111 (1810 kg ha-1 ) (susceptible) and AO-3A-ADP1-47 (218 kg ha-1 ) had the lowest yield. Interestingly, one resistant line (AO-3A-ADP1-13), had significantly higher seed yield of 1523 kg ha−1 compared to the resistant and susceptible parents (AO-1012- 29-3-3A and Rozi Koko with the seed yield of 981 kg ha−1 and 975 ha−1 respectively) AO-3A-ADP1-13 did not yield significantly different from two middle 15 American lines (PR1933-7 and PR1933-5 with the seed yield of 1187 kg ha−1 and 1236 kg ha−1 respectively). Interestingly, it was among the top three high yielding lines in Mozambique, Malawi and Zambia (Table 6).
.png)
.png)
Based on high seed yield, six lines are potential candidates either as new cultivars or improved germplasm (Table 6). These lines have the appropriate seed color belonging to the preferred southern African market classes (Figure 1). Three of the six top yielding had a similar performance for the tested traits (seed yield, days to flowering and maturity) except for seed weight as indicated in combined ANOVA across all locations (Mozambique, Malawi, US and Zambia) (Table 6).
.png)
6. Evaluation for resistance to two races of Anthracnose (Colletotrichum lindemuthianum) (Obj. 1)
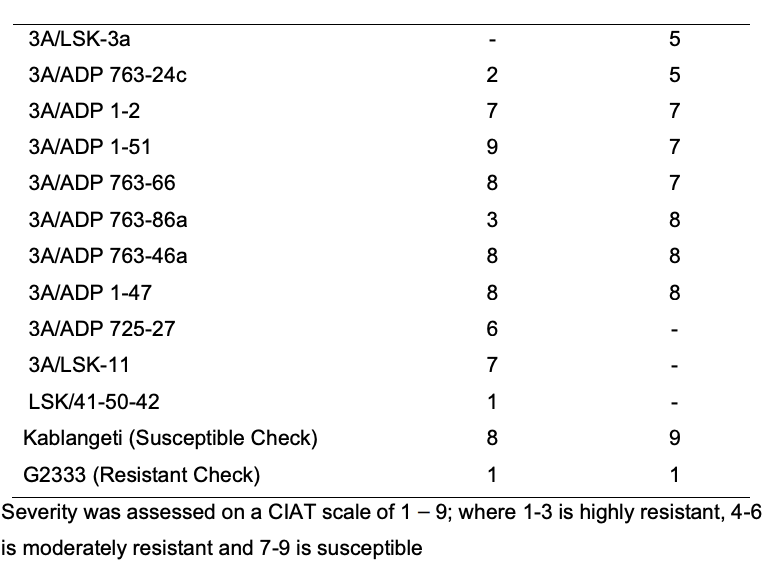
Anthracnose (ANTH) caused by C. lindemuthianum is a major disease of common bean in the three target countries for the project. The pedigree of the crosses suggested that some of the AO resistant breeding lines might have resistance to ANTH. The 16 AO resistant breeding lines were inoculated with two races of C. lindemuthianum (Races 7 and 294), which were isolated from Zambia. The artificial inoculation was conducted at UNZA greenhouses following the methodology from Mungalu et al. (2020). Four breeding lines were highly resistant to Race 7 and only one line was highly resistant to Race 294 (Table 7).
.png)
7. Search for Alternate Sources of Resistance to the APA locus (Obj. 1)
To-date, the APA locus has provided effective resistance against AO, but its resistance may start to wane and eventually fail due to selection pressure. There is need, therefore, to search for alternate sources of resistance to the APA locus, which can be pyramided with the APA locus to provide durable AO resistance. The Andean Diversity Panel (ADP), which has a rich genetic diversity of Andean germplasm, is an excellent genetic resource for identifying novel sources of resistance to the AO.
Under this project, a total of 476 ADP lines were evaluated for resistance to AO in an insectarium at the University of Zambia following the protocol described for the AO breeding lines. The data collected included the number of perforations in the seed and percentage of damaged seed. Of the 476 ADP genotypes, 51 showed damage between 0-10% (Figure 5). Even more interestingly, within this group,15 genotypes (3.2%) had zero seed damage, which is very similar to the number of 20 resistant lines found with the crosses and populations previously mentioned (2.8%). The percentage seed damage for the resistant (AO-3A) and susceptible (Kabulangeti) checks were 17.5% and 57.5%, respectively. The 15 genotypes are in variable market classes including the purple mottled (aka Kabulangeti) and the red-mottled, which are popular in southern Africa, the target region for the project. Assuming that none of the 476 genotypes in the ADP carry the APA allele from tepary bean, we hypothesize that the AO genetic resistance for these 15 genotypes may be controlled by different loci. However, additional research is needed to confirm this. The 15 genotypes that have shown higher resistance than AO-3A are currently being targeted for re-screening, and if their resistance holds, they could potentially be an important genetic resource for enhancement of common bean for AO resistance.
The ADP had already been genotyped with 32,000 SNPs from a previous project (Katuuramu et al. 2018). Taking advantage of this existing genotypic data set, an exploratory Genome-wide Association Study (GWAS) was conducted to obtain genetic insights into the basis of the observed variation in the ADP for AO resistance. Preliminary results have shown significant genomic regions associated with AO resistance in the ADP, but this is still pending confirmation in order to publish.
8. Capacity Building (Obj. 4)
The project has trained four female next-generation breeders in Plant Breeding at master's level. One was trained at NDSU (M.S. in Plant Sciences) and the other three from Malawi, Mozambique and Zambia were in the MSc. Plant Breeding and Seed System program at UNZA. This training has strengthened bean research capacity for the national bean breeding national programs in Malawi, Mozambique, and Zambia.
The other capacity building activities were through the visits of US collaborators Drs. Juan Osorno and Carlos Urrea to Mozambique and Zambia. During the project, the lead PI Dr. Juan Osorno made two trips to Zambia and one trip to Mozambique. All three trips were made when there was a bean crop in the field in both countries. Though the primary interest for the three trips were to check on the implementation of the project activities, Dr. Osorno engaged in other productive conversations with the project team members in both Mozambique and Zambia. These conversations were advisory in nature on changes or improvements that 22 can be made to respective breeding programs to enhance breeding gains. During his two visits to Zambia, Dr. Osorno also provided mentorship and input in developing study objectives for the three students studying at UNZA.
Dr. Carlos Urrea from the University of Nebraska made two trips to Zambia to support the implementation of project activities at UNZA. The first trip was to train the UNZA Bean Breeding team on how to use Field Book, an important resource in conducting breeding trials. The people who attended the training also included the bean breeder and the national bean program technician. The purpose of the second trip to Zambia that Dr. Urrea made was to install the two Mattson cookers that the project purchased and train the project team how to use them. In addition, multiple field days were made by Dr. Kelvin Kamfwa across multiple locations in Zambia. Please refer to previous annual reports for details about these activities.
Project Accomplishments
Weevil resistant breeding lines in market classes preferred in Malawi, Mozambique and Zambia have been developed and shared with the national programs in Malawi, and Mozambique. These breeding lines are currently being evaluated for agronomic performance in the three target countries.
The 16 AO-resistant breeding lines were evaluated for cooking time using the Mattson cooker. Breeding lines with the ideal combination of weevil resistance and faster cooking time were identified.
The 16 AO-resistant lines were evaluated for agronomic performance in preliminary yield trials. Weevil-resistant breeding lines with higher productivity were identified.
An InDel marker (α-AI) was validated for use in MAS. This marker showed 100% prediction accuracy for the weevil resistance phenotype and can be used in MAS 23 for weevil resistance to circumvent the lengthy period required for phenotyping for AO resistance.
The project has also strengthened the University of Zambia research capacity to conduct lab-based cooking time trials using the Mattson cookers. The project purchased and installed two Mattson cookers for the breeding program at UNZA. These cookers will be used in routine screening for cooking time and will provide a service to other bean research groups in southern Africa who want to conduct cooking time experiments on common bean.
The project has trained four female next-generation breeders in Plant Breeding at Master level. One was trained at NDSU, and the other three from Malawi, Mozambique, and Zambia were in the MSc. Plant Breeding and Seed System program at UNZA. This training has strengthened bean research capacity for the national bean breeding national programs in Malawi, Mozambique, and Zambia.
Furthermore, several undergraduates, research interns, and technicians in Malawi, Mozambique, and Zambia have been trained in conducting weevil resistance, cooking time, and field trials. This training will be critical to sustaining breeding efforts for weevil resistance in breeding programs in the three target countries.
The project also provided an opportunity for the UNZA breeding program to meaningfully collaborate with the private sector, specifically Good Nature Agro (GNA), to ensure that the weevil-resistant varieties developed by the project are scaled-up in production and distribution to farmers.
Highlights of Addressing Cross-Cutting Themes (HICD, Gender, Youth, Nutrition, Resilience)
Gender was mainstreamed in the project at various levels. The project organization's Co-PI’s from Malawi and Mozambique are female. In capacity building, the four students from Malawi, Mozambique, and Zambia that the project 24 has trained at Master level as next generation bean breeders are all female. Among the four interns that have been working on the project in Zambia, two are female. These four interns are youths that are below 30 years old.
Utilization of Research Outputs and Handoff/Scaling of the Outputs
The 16 AO-resistant genotypes have been shared with the national bean breeding programs in Malawi, Mozambique, and Zambia, where they have been evaluated for agronomic performance. After this evaluation, breeders in each of these countries will make decisions on the line/s to be moved forward for commercial release as cultivars. The model that will be used for seed dissemination and scaling-up production will depend on each of the three countries seed regulations and policies. In the case of Zambia, the 16 breeding lines will be tested in preliminary yield trials (PYT), and the top performing lines will then be tested in advanced yield trials (AYT) and on-farm trials. It is after these trials that a decision will be made on the breeding lines to be submitted to Seed Control and Certification Institute (SCCI), which is a government agency that regulates the regulates the seed industry, including release of varieties. In PYT, AYT, and on-farm trials, GNA will continue to be involved as a private sector partner. GNA will provide testing sites for on-station trials and on-farm testing sites of smallholder farmers that they have been working with to produce common bean. Once the weevil resistant varieties are released, GNA will be the suitable partner for scaling-up and handing over of the improved varieties subject to licensing agreements. These agreements, including royalties will ensure that some of the revenue from these AO-resistant varieties can be used to support the UNZA Bean Breeding program sustainably.
In addition to GNA, UNZA Breeding Program has lead farmers in major bean producing districts of Zambia that were previously important in disseminating and adopting improved varieties developed by the program. These farmers will again be used in on-farm trialing of AO-resistant breeding lines and subsequent scalingup of production of released varieties.
Further Challenges and Opportunities
The major challenge the project has experienced is the low recovery of resistant genotypes from the crosses. Of the first set of 574 breeding lines from AO-3A, only 16 (2.8%) were resistant. Similar results have been found in the screening of the second set and the ADP. The implication of this result is that breeding programs seeking to breed for weevil resistance should work with sufficiently large breeding population sizes to increase the chances of identifying resistant lines.
The project has demonstrated that the APA locus is stable in many genetic backgrounds. However, its effectiveness depends, to some extent, on the genetic background. The 16 resistant lines containing the APA locus showed variable resistance levels, with some having even higher resistance levels than the resistant parent AO-3A. The behavior of the APA locus in different genetic backgrounds presents an opportunity to develop resistant genotypes with higher resistance levels than that of the existing resistant genotypes.
The molecular marker that the project has validated opens up opportunities to enhance/facilitate and speed up breeding efforts for weevil resistance through marker-assisted selection.
The project has provided a breeding solution to a major problem of post-harvest losses in common bean in Malawi, Mozambique, and Zambia. The AO-resistant breeding lines that the project has developed present an opportunity to enhance food security in the three target countries.
References
Kusolwa, P.M., J.R. Myers, T.G. Porch, Y. Trukhina, A. González-Velez, and J.S. Beaver. 2016. Registration of AO-1012-29-3-3A red kidney bean germplasm line with bean weevil, BCMV and BCMNV resistance. J. Plant Regist. 10:149–153.
Cichy, K., T. Porch, J. Beaver, P. Cregan, D. Fourie, R. Glahn, M. Grusak, K. Kamfwa, D. Katuuramu, and P. McClean. 2015. A Phaseolus vulgaris diversity panel for Andean bean improvement. Crop Sci. 55:2149–2160.
Mazaheri, L. I. (2018). Development of a Molecular Marker to Track APA G40199 Introgression in Common Bean for Bruchid Resistance (Master’s Thesis North Dakota State University).
Mungalu, H., Sansala, M., Hamabwe, S., Mukuma, C., Gepts, P., Kelly, J. D., and Kamfwa, K. 2020. Identification of race-specific quantitative trait loci for resistance to Colletotrichum lindemuthianum in an Andean population of common bean. Crop Science 60(6): 2843–2856.
Katuuramu, D. N., J.P. Hart, T.G. Porch, M.A. Grusak, R.P. Glahn, and K.A Cichy. 2018. Genome‐wide association analysis of nutritional composition‐related traits and iron bioavailability in cooked dry beans (Phaseolus vulgaris L.). Molecular Breeding 38(4): 44.



 Print
Print Email
Email



